Legumes are valued worldwide as a sustainable and inexpensive food source and are considered second only to cereals for global food security 1. In addition to their nitrogen-fixing ability, they are nutritionally valuable providing proteins, essential amino acids, complex carbohydrates and dietary fiber, vitamins and minerals 1, 2 . They have also been ascribed cultural and medicinal roles due to their possession of active bio-compounds 1. For thousands of years this cultural significance has influenced our selective breeding of only the most palatable and fecund legumes 3. It has followed that in modern times the focus of our scientific research has remained limited to this exclusive, select few. Within the field of genomics, this has given rise to a group of ‘model’ legumes from which all of our current genetic understanding is based.

Rapid developments in DNA and RNA sequencing have greatly advanced our genomic knowledge of these model legumes. Their reference genomes have allowed researchers to decipher key genes, pathways, and networks regulating biological mechanisms and important agronomic traits 2, 4. This has allowed for more targeted and efficient breeding and cross-breeding of some major economic legumes such as the Peanut 5.
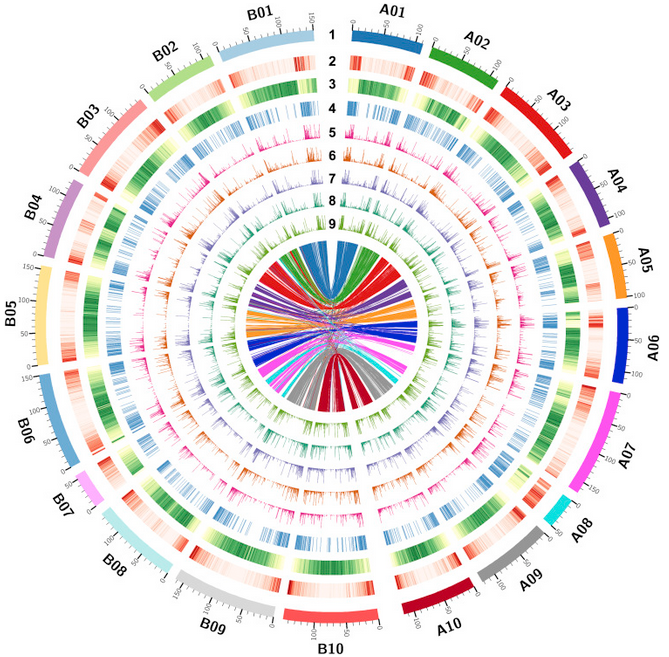
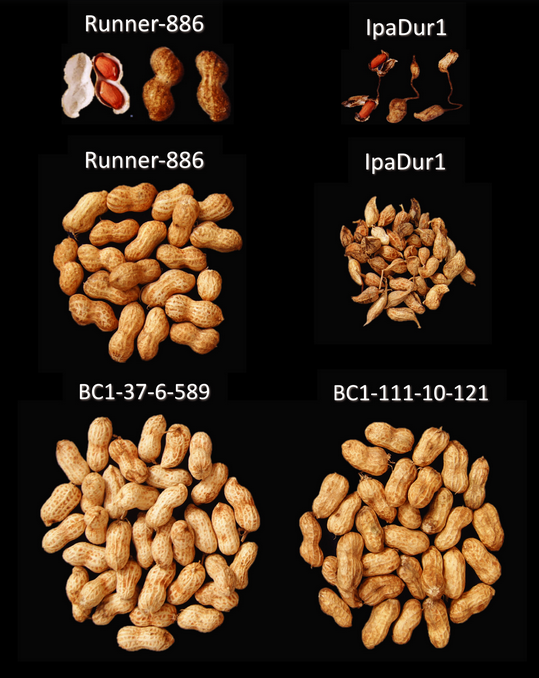
While this is undoubtedly crucial knowledge for food security, our genomic maps remain largely limited to sphere of dietary legumes. The rest is uncharted territory. We have little genomic knowledge of forage legumes in comparison, leaving our agricultural systems vulnerable to the effects of climate change. But, thanks to organizations like the International Center for Tropical Agriculture (CIAT), we do now have genomic references for some of the major animal forage and fodder legumes, including Alfalfa, Soybean and Cowpea 6.
The Cowpea (Vigna unguiculata) for example is one of the most important forage legumes in tropical and sub-tropical regions with over 14 million hectares of agricultural land planted with it across the globe 7. Thanks to CIAT’s Tropical Legumes Projects, we have identified locations within its genomic map that code for important agronomic traits including drought resistance and efficient seed development.

Genetic markers have been used to target these locations and facilitate more efficient and directed breeding depending on what producers need – (I discuss in the previous post how we might better understand ‘what producers need’ through a conceptual approach that incorporates the different ago-ecological dimensions of a farming system).

Despite these major breakthroughs, large gaps remain in the forage legume germplasm collection. It is still a long way from from being representative of the total geographic diversity of tropical legumes 8.
Yet, in a round-about way, this is a positive as it can mean only one thing – that there is an unknown quantity of functionally important genes out there in the wilderness, just waiting to be discovered. Within this diversity are almost certainly to be unique sequences not found within our model set that confer novel ways to adapt and survive the growing effects of climate change. Through the continued genomic mapping of these wild and geographically dispersed species, we can begin to chart new territory. We can learn how these genes are linked to others and how they are passed down through generations. From there we can learn how to breed them quickly and efficiently and integrate them into our agricultural systems.
To keep with the vernacular, what Mr. Peanut and his exclusive club of model legumes need are a few wild characters to liven up the party!
1 -Maphosa, Y., & Jideani, V. A. (2017). The role of legumes in human nutrition. Functional food-improve health through adequate food, 1, 13.
2 – Dai, X., Zhuang, Z., Boschiero, C., Dong, Y., & Zhao, P. X. (2021). LegumeIP V3: from models to crops—an integrative gene discovery platform for translational genomics in legumes. Nucleic Acids Research, 49(D1), D1472-D1479.
3- Liu, J., Yu, X., Qin, Q., Dinkins, R. D., & Zhu, H. (2020). The impacts of domestication and breeding on nitrogen fixation symbiosis in legumes. Frontiers in genetics, 11, 973.
4 – Pandey, M. K., Roorkiwal, M., Singh, V. K., Ramalingam, A., Kudapa, H., Thudi, M., … & Varshney, R. K. (2016). Emerging genomic tools for legume breeding: current status and future prospects. Frontiers in Plant Science, 7, 455.
5 -Vishwakarma, M. K., Nayak, S. N., Guo, B., Wan, L., Liao, B., Varshney, R. K., & Pandey, M. K. (2017). Classical and molecular approaches for mapping of genes and quantitative trait loci in peanut. In The peanut genome (pp. 93-116). Springer, Cham.
6 -Kulkarni, K. P., Tayade, R., Asekova, S., Song, J. T., Shannon, J. G., & Lee, J. D. (2018). Harnessing the potential of forage legumes, alfalfa, soybean, and cowpea for sustainable agriculture and global food security. Frontiers in plant science, 9, 1314.
7-Agbicodo, E. M., Fatokun, C. A., Muranaka, S., & Visser, R. G. (2009). Breeding drought tolerant cowpea: constraints, accomplishments, and future prospects. Euphytica, 167(3), 353-370.
8-Schultze-Kraft, R., Peters, M., & Wenzl, P. (2020). A historical appraisal of the tropical forages collection conserved at CIAT. In Genetic Resources.
