We are trying to resolve the issue of the dye staining the glass, once I get these specs removed I will re do this PCR with a higher concentration of DNA

We are trying to resolve the issue of the dye staining the glass, once I get these specs removed I will re do this PCR with a higher concentration of DNA
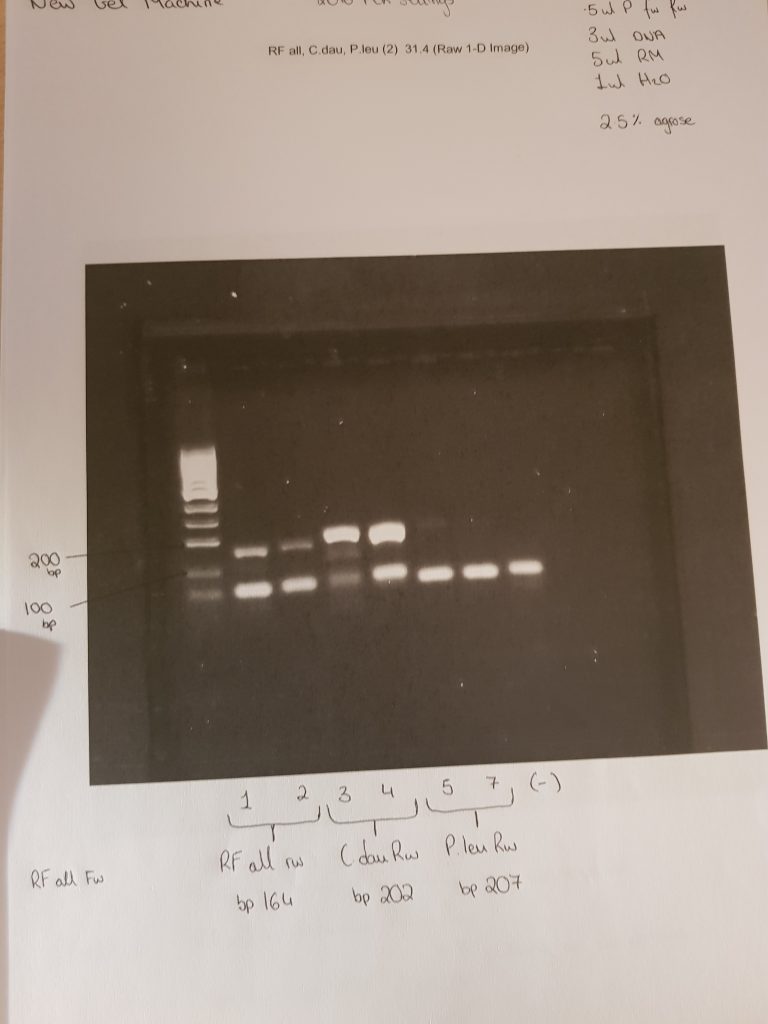

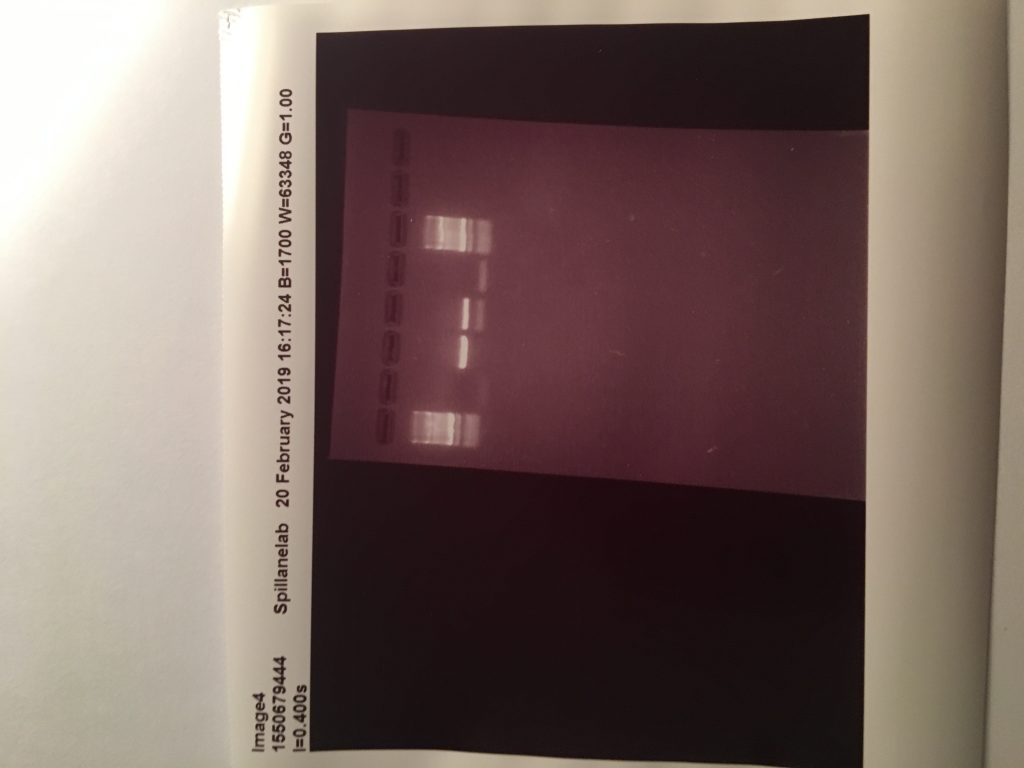
These two images show positive results for Rumen fluke from individually isolated eggs from faecal samples and using the ITS2 Flks primers and PCR settings from the 2016 paper. These results mean I have sufficient amounts of positive DNA pool of rumen fluke to begin my samples from the better farm project.
My next step is trying to get my species specific Primers to work by figuring out their specific PCR settings. My course coordinator designed species specific primers for me, however, I need to know the PCR settings for them that will work to continue.
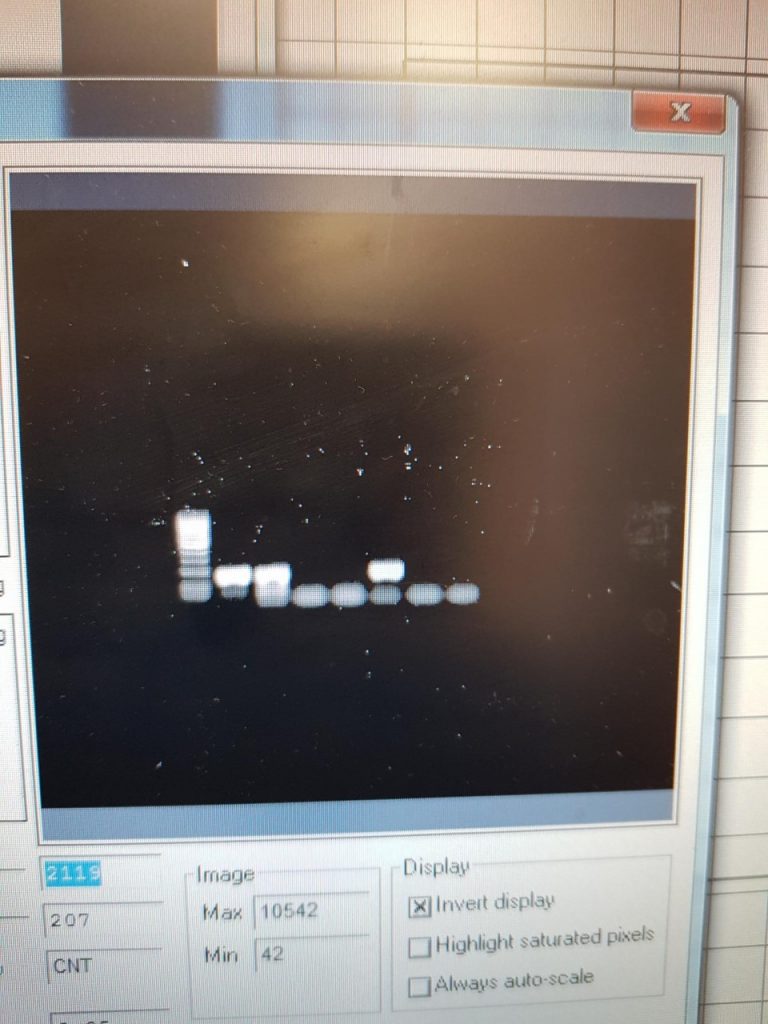
This PCR was carried out on 18/4/19
The first two samples were 100 isolated eggs, stored overnight in ethanol, and broken with a sonicbeater. Then using the QIAamp stool kit I washed and eluted the sample and recorded the DNA concentration using a nanodrop. The other 5 samples are from faecal samples of cattle I collected on the teagasc farm. The primers used were the ITS2 FLKS from the 2016 paper.
This PCR now shows ive working primers and PCR settings.
In previous weeks I was carrying out DNA extraction, PCR and gel electrophoresis, these tests were giving me negative results.
Firstly I was not getting sufficient levels of DNA from my faecal samples, to solve this problem I cleaned the faecal samples from the animals by sedimentation and cleaning the samples down till just the eggs and very small amounts of debris were present. Using a microscope and a Pasteur pipette i individually isolated the eggs from this sample and recorded the exact number of eggs present. This technique ensured that I had only Rumen Fluke DNA resent in my sample. The next step was making sure that I was successfully breaking up the eggs so that the DNA was released, this took me over 2 weeks as I was trying different methods such as freeze thaw cycles, boiling, soaking in ethanol, the stool kit, the tissue kit and a hand homogeniser. Eventually we succeeded on breaking up the eggs to relieve the DNA by soaking the sample over night in ethanol and using a sonic beater to use sound waves to break the egg.
The next problem I was having which I have resolved is using the correct PCR settings. My supervisor and I designed primers previously in NUIG, and for these primers to work I would have required different settings for my PCR. Once I ensured I was able to extract the DNA successfully from the rumen fluke eggs i asked my supervisor to order primers from a 2016 research paper on the ITS2 region of rumen fluke, where they also published the settings for us to follow exactly. (However they do not specify the base pair length, so I need to find out the bp length for the 2016 paper primers and the PCR settings that will work with the previously made primers by Galina for me in college )
Last week was a very busy week for me in the laboratory and on the farms.
Monday was spent screening faecal samples from blackface mountain ewes from county Galway. These ewes contained high amounts of both rumen and liver fluke, this is understandable as they come from a wet area which could give rise to the intermediate host, and the owners of mountain sheep would be less likely to regular dose their sheep against fluke.
Tuesday was my first day collecting samples on a farm. Michael Fagan who is the farm manager brought me two Teagasc farms Tuohy’s and Newford, Tuohy’s is a rented farm where they rear their steer (castrated) bull claves for dawn meats and Newford is where they have their suckler herd. Michael brought me out to take samples from Tuohy’s because he felt that these bullocks were not thriving as well as before Christmas and suspected a fluke infection. I collected 11 samples, six from tagged calves and five from ground samples.
After Tuohy’s we headed for Newford where I collected 18 faecal samples from cows around the farm. One cow on this farm was quite ill so samples had previously been taken from this cow and both rumen and liver fluke eggs were identified. I collected samples from this cow and 16 others in the hope of identifying rumen fluke eggs.
These 29 samples were stored in the fridge overnight. On Wednesday and Thursday, I prepared both Tuohy’s and Newford samples through the sedimentation method and the addition of the methylene blue and began to screen the slides.
On Friday I continued to screen these samples and recorded rumen fluke eggs in only one of the samples from the Tuohy farm, next week I will run PCR on all 11 samples to see if there is rumen fluke DNA in the samples. If there is RF DNA in the samples where no eggs were found, then the animal is infected with the immature stage which is causing harm to the animal but not shedding eggs.
Next week I will continue screening, running PCR and finish the corrections for my Lit review.
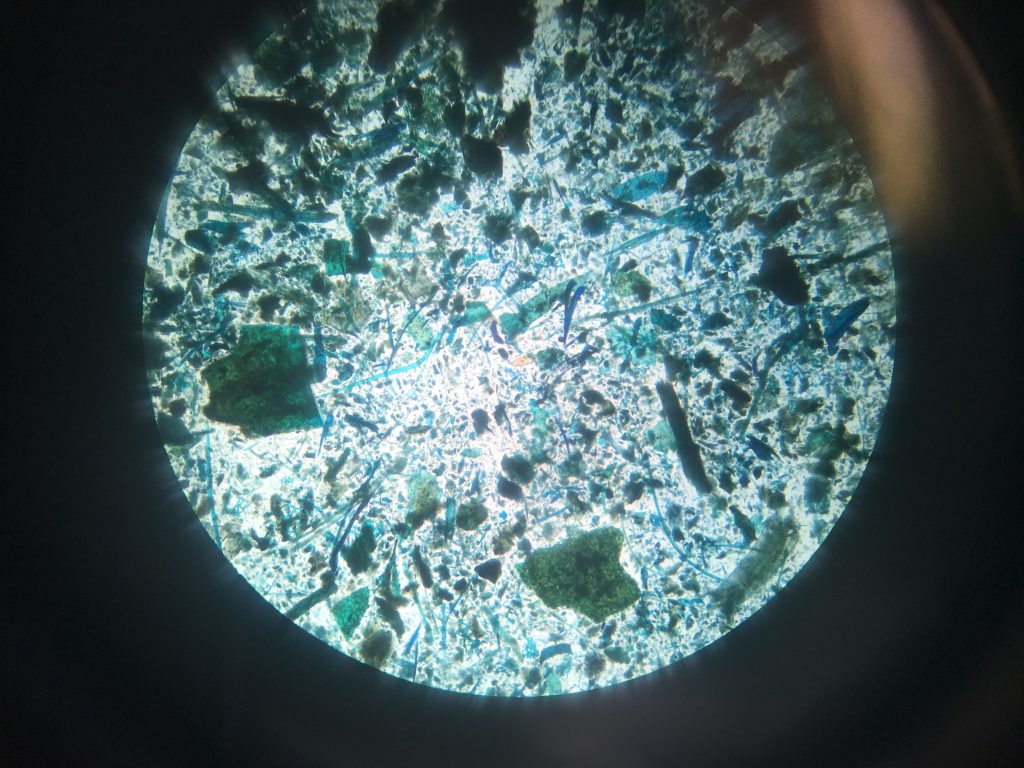



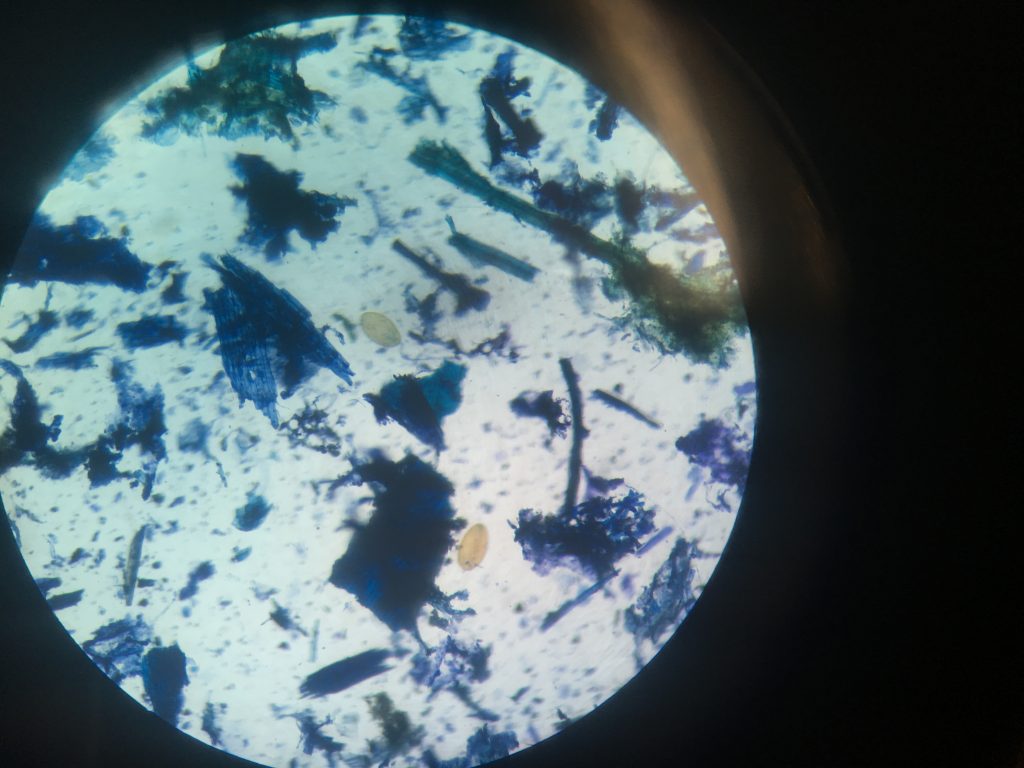
Fluke egg identification under the microscope. This faecal sample was taken from sheep in Leenane Co.Galway, I will run PCR to see if this animal contains both Liver and Rumen fluke.
My week was mostly spent in NUIG with my course coordinator Galina and my lab partner, here I learned how to carry out extraction of DNA from a Haemonchus contortus. This is the parasite my partner will be carrying out his master’s project on. Post extraction I learned how to perform washing and elution of the DNA extracted from the worm and then we performed PCR.
This was my first time to carry out this laboratory work and it worked successfully, we were also shown how to perform qPCR on our samples. However, we could not make a quantitative PCR as we do not have other samples of DNA present to compare the levels in the sample, because we extracted the DNA form the worm and not the eggs of the sample.